TU Dortmund Researchers Report an Advance in DNA Analytics
- Research
- Top News
- Press Releases
DNA – the cell’s genetic material – is made up of four building blocks, each containing one of the DNA bases: A, G, C, or T. These building blocks are joined in a long strand that forms the DNA double helix through ordered pairing with the DNA bases of a second strand (A pairs with T, G with C). In the human genome, the DNA base C can be modified differently when it occurs in the base sequence CG. This sequence and its exact opposite occur in the two strands of the double helix, and the C contained in both strands can also be modified.
These modifications play a significant role in controlling which genes in a cell are switched on or off. Thus, they are involved in forming the large number of different cell types in the human body, such as nerve cells or white blood cells. In addition, it has long been known that abnormally inserted or missing modifications of DNA building blocks can disrupt cell growth and transform a healthy cell into a proliferating cancer cell. For this reason, scientists are trying to find out exactly where these modifications occur.
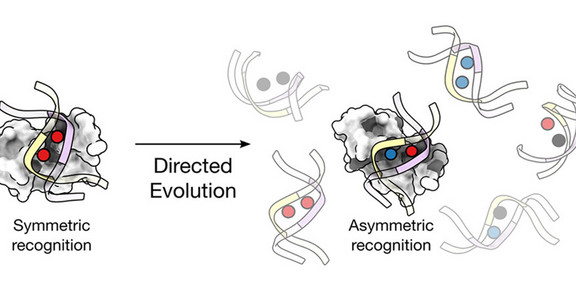
Simultaneous analysis of both DNA strands
While all four possible modifications and their exact positions can already be routinely detected in DNA in a strand-specific manner, it was not previously possible to do this simultaneously for both strands of an individual DNA double helix. Now, for the first time, Dr. Benjamin Buchmuller from Prof. Daniel Summerer's research group in the Department of Chemistry and Chemical Biology has created protein probes, using so-called directed evolution, which can selectively recognize a specific combination of two different modifications in both strands of the CG sequence.
Initial structural insights into how this type of probe recognizes DNA were gained through a collaboration with Prof. Rasmus Linser. In addition, the first functional tests showed that the probe can be used to detect small amounts of DNA with the corresponding double helix modification in the presence of larger amounts of human genomic DNA.
Together with researchers at the Chemical Genomics Center of the Max Planck Institute for Physiology in Dortmund, the researchers are now trying to use the probe to create more precise genomic maps of this double helix modification, for example to gain new insights into embryonic development and to find biomarkers for cancer.
The study published in the Journal of the American Chemical Society was carried out as part of the project EPICODE. For this project, Prof. Daniel Summerer has received nearly two million euros in funding from an ERC Consolidator Grant awarded by the European Research Council in 2017. Prof. Malte Drescher’s group at the University of Konstanz also contributed to the studies.
Contact for further information:




![[Translate to English:] Partner Four hands are holding the green logo of TU Dortmund University](/storages/tu_website/_processed_/1/d/csm_Partner_Nicole_Rechmann_KW_670eba0154.jpg)




![[Translate to English:] Forschung An apparatus with tubes in a laboratory](/storages/tu_website/_processed_/0/c/csm_Forschung_Juergen_Huhn_4fa3153b51.jpg)
![[Translate to English:] Studium Five students are sitting in a lecture hall. They are talking to each other.](/storages/tu_website/_processed_/c/9/csm_Studium_FelixSchmale_dbdbfb0dd7.jpg)





