Chemical Toolbox Offers New Insights Into Proteins
- Top News
- Research
The Emmy Noether Independent Junior Research Group led by Dr. Malte Gersch from the Department of Chemistry and Chemical Biology has developed a chemical toolbox that provides insights into how the special protein fubi works. The scientists recently published their findings in the renowned journal Nature Chemical Biology.
The small protein ubiquitin is particularly well-known for marking other proteins for their degradation, and thus helps regulate all cellular processes. This marking prevents, for example, proteins that are faulty or no longer required from accumulating in the cells. Various other ubiquitin-like proteins exist alongside the ubiquitin system, including fubi, which has not been thoroughly studied despite its immunomodulatory activity. Fubi is made in fusion with the ribosomal protein S30. In order for the ribosomes – the cell's protein factories – to function, fubi has to be separated from S30.
Insights into how the protein works
Immune cells use this by-product of ribosome production as a signal protein, for example to locally reduce the activity of the maternal immune system in the uterus, thus enabling embryos to implant. How fubi is specifically recognized by proteases for cleaving S30, and how it differs from ubiquitin was previously unknown.
Scientists led by Dr. Malte Gersch, who heads the Emmy Noether Independent Junior Research Group based at the Department of Chemistry and Chemical Biology and at the Chemical Genomics Center of the Max Planck Institute for Molecular Physiology, has now gained initial molecular insights into the mechanism that facilitates the fubi-controlled ripening of the key protein S30.
“Toolbox“ promotes understanding of the fubi system
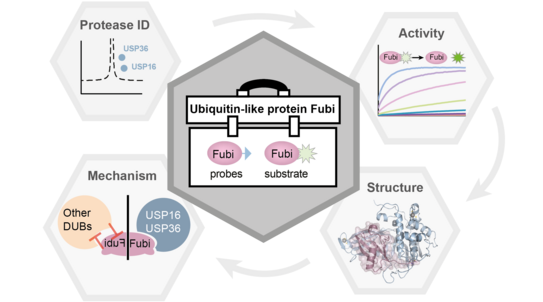
Using a newly developed chemical toolbox, the researchers were able to determine at the molecular level how the two deubiquitinating enzymes USP16 and USP36 have another specific fubi protease activity. This double fubi/ubiquitin specificity allows fubi-S30 maturation in two spatially determined environments of the cell, while also preventing the large number of other only ubiquitin-specific enzymes from processing fubi.
“Our results show how molecularly entangled the ubiquitin system is with the previously underresearched fubi system. Combined with the chemical-biological toolbox we have developed, this will also expand the understanding of fubi as a protein modification in cellular processes,” says Dr. Malte Gersch.
Contact person for queries:




![[Translate to English:] Partner Four hands are holding the green logo of TU Dortmund University](/storages/tu_website/_processed_/1/d/csm_Partner_Nicole_Rechmann_KW_40b35bb3fd.jpg)




![[Translate to English:] Forschung An apparatus with tubes in a laboratory](/storages/tu_website/_processed_/0/c/csm_Forschung_Juergen_Huhn_cbd34afd6d.jpg)
![[Translate to English:] Studium Five students are sitting in a lecture hall. They are talking to each other.](/storages/tu_website/_processed_/c/9/csm_Studium_FelixSchmale_81d94adc86.jpg)





