It's All About Dynamics
- mundo

To understand how proteins work, decoding their structure is not enough. Knowing how they move and interact with their environment is equally important. In collaboration with international partners, Prof. Rasmus Linser from the Department of Chemistry and Chemical Biology is further developing a method called NMR spectroscopy, which is capable of observing the building blocks of life in action.
Proteins carry out a variety of tasks in our bodies. They give our cells structure and allow them to move, transport metabolic products, determine our responses to diseases and, as enzymes, catalyze numerous biochemical reactions. They are always on the move. Because they can only carry out these tasks by flexibly changing their shape – for instance, to identify certain target structures or to bind to reactants.
“If we want to understand how proteins work, these dynamics are essential,” says Prof. Rasmus Linser from the Department of Chemistry and Chemical Biology. “However, observing proteins in motion is a major challenge.” Movements are often a hindrance to decoding the three-dimensional structure of proteins. For traditional methods of structural analysis, the proteins under examination are therefore usually examined at extremely low temperatures in a crystal lattice. Although this approach allows the protein structure to be displayed in high resolution, their dynamics remain hidden.
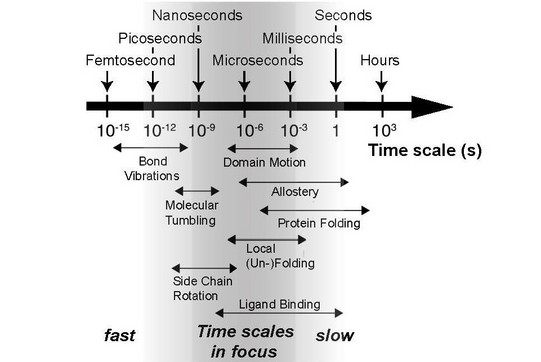
That is why Linser works with a different technique. Nuclear magnetic resonance spectroscopy – or NMR spectroscopy for short – makes it possible to record not only the structure of proteins in a sample but also the dynamics. “The basic physical principle is the same as with MRI, which is used in medicine to take pictures of bones and organs, but with different technical facets,” explains Linser. A sample is exposed to complex sequences of electromagnetic pulses within a strong magnetic field. It is then possible to use the resulting spectra to deduce the distances between the individual atomic nuclei, how they are embedded in the protein structure, and how they interact with neighboring atoms. In addition to that, movements can be recorded on different time scales. The dynamics in the range from picoseconds to milliseconds, in which the diverse mechanisms of protein functionality often occur, are of particular interest here.
Exploring and expanding boundaries
Linser's research group has several large NMR devices that generate a strong magnetic field and can analyze samples under different conditions. Depending on the application, the proteins are either in solution or examined as solids. “One of the key limitations so far is that NMR spectroscopy is only suitable for relatively small proteins,” says Linser. Because the more complex a protein is, the more confusing the spectrum becomes, at which point evaluation is no longer possible, even with specialized computer programs. “In our research, we explore the limits of what is possible and try to overcome them more and more using innovative approaches,” says Linser.
The TU professor receives financial support from the European Research Council (ERC), which selected Linser’s project for one of the prestigious Consolidator Grants in early 2023 and will provide it with around two million euros in funding over the next five years. “We combine various approaches to get a better handle on the complexity,” Linser explains. “For a start, we vary the experimental approaches – for instance, the chemical nature of the sample, the use of hardware tailored to it and in particular the patterns according to which magnetization is transferred back and forth between atomic nuclei and coded differently, known as pulse sequences. We also start by analyzing the raw data and then develop new approaches to data reconstruction and utilization.” Linser is always careful to link methodological advances directly with practical applications. “We are collaborating with various research groups, both within TU Dortmund University and worldwide,” he explains.
International collaboration
One of the largest proteins that Linser’s team has analyzed so far using NMR spectroscopy as part of an international collaboration is a modified form of tryptophan synthase. This protein is found in bacteria, among other things, and is of interest for both medical research and biotechnology. “Research teams from California and Wisconsin, with whom we collaborate, have used artificial evolution to develop a variant of the naturally occurring protein that can be used far more effectively for industrial processes,” explains Linser. "But the question is: Why does this work? The biggest differences are usually not in the structure, but in the dynamics. And that’s where we hope to make a contribution.”
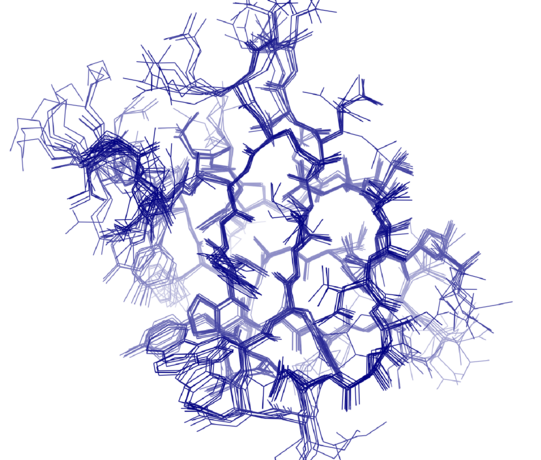
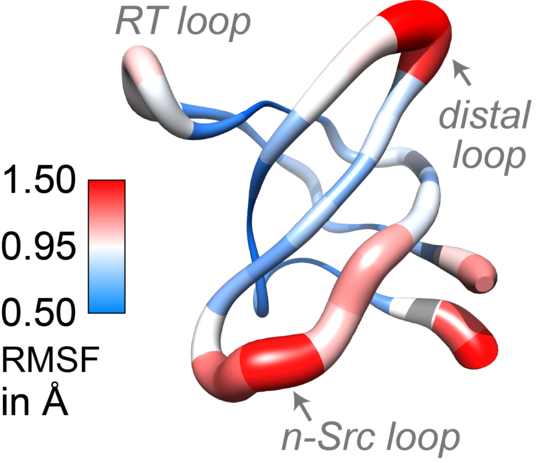
So far, NMR spectroscopy has already reached its limits with proteins with a molecular mass of 40 kilodaltons. Tryptophan synthase, on the other hand, consists of two subunits with a mass of 72 kilodaltons each – a welcome challenge for Linser. In fact, with the help of new experimental approaches and innovative data processing, his team managed to create the basis for explaining dynamic processes within these proteins.
Linser's team also works in the RESOLV Cluster of Excellence, which is coordinated by TU Dortmund University and Ruhr-Universität Bochum as host universities. “RESOLV is about the role that the solvent plays in various processes, for example in catalysis, in an electrochemical or in a biochemical context,” explains Linser. “I’m particularly interested in how proteins interact with it and use it.”
In this context, he focused on the human enzyme carbonic anhydrase. Carbonic anhydrase catalyzes the conversion of water and carbon dioxide into carbonic acid and vice versa. It plays an important role in our breathing and in acid regulation in the stomach and kidneys, among other things. The active center, i.e. the reaction pocket in which the conversion takes place, is key to how carbonic anhydrase functions.

“With the help of NMR spectroscopy, we have demonstrated that the active center undergoes a conformational movement that also influences the ‘permitted’ positions of the water molecules in the pocket,” says Linser. Previously, carbonic anhydrase had mainly been studied using methods that omit this dynamic. It was only after Linser and his team developed the options of NMR spectroscopy that it became possible to use this technique to analyze the protein, which is also relatively large at around 30 kilodaltons, and record movements on a time scale in the microsecond range. It was proven for the first time that the protein can imprint varying properties onto the water around the active center through its strong interaction with the water in the pocket – despite the high mobility of the individual solvent molecules.
Focusing on solvents
“We are currently researching how dynamic processes interact in different elements of the pocket. In doing so, we also include small, mostly synthetic molecules, known as ligands, which bind to the active center,” explains Linser. “The water is the connecting element. We want to understand how exactly the unity of solvent and protein affects the function.” He already has ideas for the upcoming phase of the Excellence Strategy starting in 2026. If the RESOLV Cluster of Excellence is extended, he could easily imagine researching how the internal dynamics of ligands, which bind to proteins with very different functions, are impacted by different solvents.
„We want to link methodological advances directly with practical applications. To achieve this, we collaborate with research groups worldwide.“ Prof. Rasmus Linser
“Ligands of this kind can activate, inhibit or otherwise modulate the function of their respective target protein. Or they are converted into new products as substrates by the respective protein,” explains Linser. “In a biochemical context, the solvent probably influences the interaction in more diverse ways than previously assumed. It could make larger, more complex ligands and substrates more flexible and facilitate the process of binding to the protein, or even complicate the process.” When it comes to the biotechnological synthesis of larger molecules in particular, such information would be extremely helpful in making the respective reactions as effective as possible.
“So far, it has been very difficult to predict how a specific solvent will influence molecular dynamics,” says Linser. “Although the solvent is ubiquitous, detecting it through spatially resolved experiments is challenging.” The solvent is usually not observed directly via NMR spectroscopy either – but its influence on the movement of the proteins and their binding partners is. “Developing this field further as part of RESOLV is definitely an attractive prospect,” said Linser. "With our equipment and expertise, we are in an ideal position to advance research in this field as well."
Text: Elena Bernard
About:

Prof. Dr. Rasmus Linser has been a Professor of Physical Chemistry at the Department of Chemistry and Chemical Biology since 2018. He studied chemistry in Göttingen and Madrid and received his doctorate in 2010 from the Leibniz Institute for Molecular Pharmacology in Berlin. For four years, he researched on a rotating basis at the University of New South Wales in Sydney, Australia, at Harvard Medical School in Boston, USA, and at the Walter and Eliza Hall Institute in Melbourne, Australia. From 2014, he headed an Emmy Noether independent junior research group at the Max Planck Institute for Biophysical Chemistry in Göttingen, and from 2016 to 2018, he was a professor at Ludwig-Maximilians-Universität München. At TU Dortmund University, he is researching improvements in NMR spectroscopy to shed light on the structure, dynamics and function of proteins.
This is an article from mundo, the research magazine of TU Dortmund University.




![[Translate to English:] Partner Four hands are holding the green logo of TU Dortmund University](/storages/tu_website/_processed_/1/d/csm_Partner_Nicole_Rechmann_KW_40b35bb3fd.jpg)




![[Translate to English:] Forschung An apparatus with tubes in a laboratory](/storages/tu_website/_processed_/0/c/csm_Forschung_Juergen_Huhn_cbd34afd6d.jpg)
![[Translate to English:] Studium Five students are sitting in a lecture hall. They are talking to each other.](/storages/tu_website/_processed_/c/9/csm_Studium_FelixSchmale_81d94adc86.jpg)





